
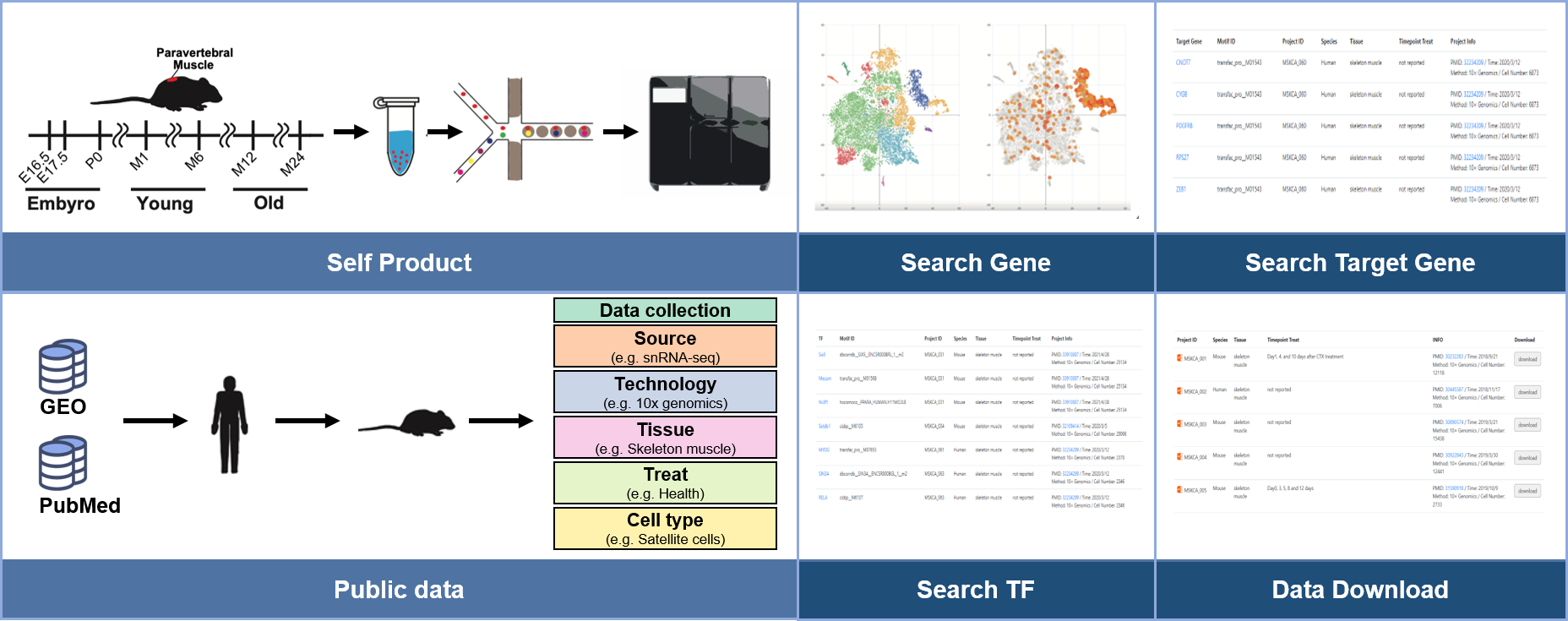
To uncover the role of satellite cells (SCs) in paravertebral muscle development and aging, we constructed a single-nucleus transcriptomic atlas of mouse paravertebral muscle across seven timepoints spanning the embryo (day 16.5) to old (month 24) stages. Eight cell types, including SCs, fast muscle cells, and slow muscle cells, were identified. An energy metabolism-related gene set, TCA CYCLE IN SENESCENCE, was enriched in SCs. Forty-two skeletal muscle disease-related genes were highly expressed in SCs and exhibited similar expression patterns. Among them, Pdha1 was the core gene in the TCA CYCLE IN SENESCENCE; Pgam2 , Sod1 , and Suclg1 are transcription factors closely associated with skeletal muscle energy metabolism. Transcription factor enrichment analysis of the 42 genes revealed that Myod1 and Mef2a were also highly expressed in SCs, regulate Pdha1 expression, and are associated with skeletal muscle development. These findings hint that energy metabolism may be pivotal in SC development and aging. Three ligand-receptor pairs of extracellular matrix (ECM)-receptor interactions: Lamc1-Dag1 (laminin subunit gamma 1–dystroglycan), laminin subunit alpha 2 ( Lama2 )- Dag1 , and heparan sulfate proteoglycan 2 ( Hspg2 )- Dag1 , may play a vital role in SC interactions with slow/fast muscle cells and SC self-renewal. Finally, we built the first database of a skeletal muscle single-cell transcriptome: the Musculoskeletal Cell Atlas (http://www.Skeletalmuscle.tech), which lists 630,040 skeletal muscle cells and provides interactive visualization, a useful resource for revealing skeletal muscle cellular heterogeneity during development and aging. Our study could provide new targets and ideas for developing drugs to inhibit skeletal muscle aging and treat skeletal muscle diseases.